Individual Based Simulation of the accumulation of junctions
Source:R/sim_inf_chrom.R
sim_inf_chrom.RdIndividual based simulation of the accumulation of junctions for a chromosome with an infinite number of recombination sites.
Usage
sim_inf_chrom(
pop_size = 100,
freq_ancestor_1 = 0.5,
total_runtime = 100,
morgan = 1,
markers = -1,
seed = 42
)Arguments
- pop_size
Population Size
- freq_ancestor_1
Frequency of ancestor 1 at t = 0
- total_runtime
Maximum time after which the simulation is to be stopped
- morgan
Mean number of crossovers per meiosis (e.g. size in Morgan of the chromosome)
- markers
The number of genetic markers superimposed on the chromosome. If markers is set to -1, no markers are superimposed (faster simulation)
- seed
Seed of the pseudo-random number generator
Examples
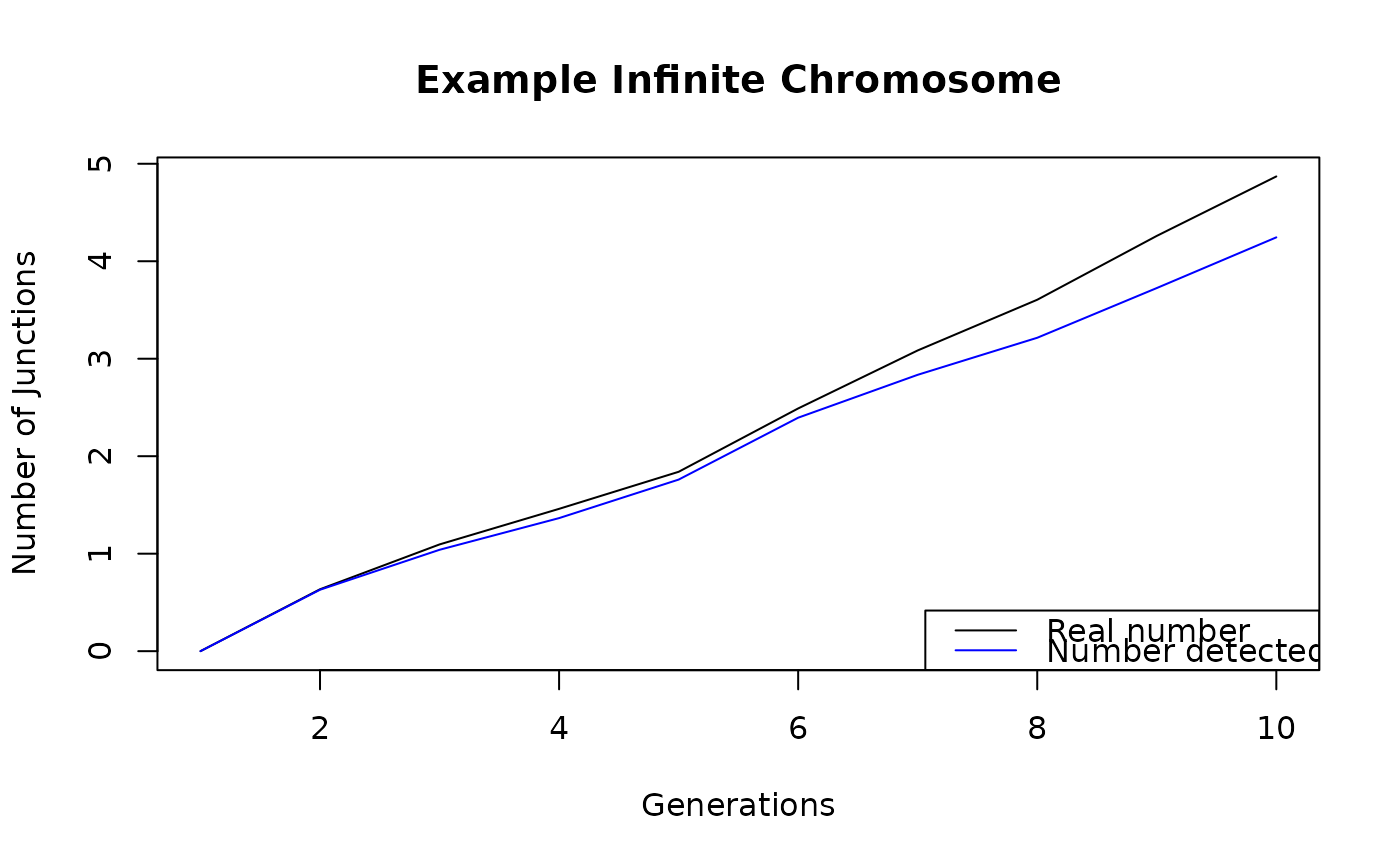
v <- sim_inf_chrom(pop_size = 100, freq_ancestor_1 = 0.5,
total_runtime = 10, morgan = 1, markers = 100,
seed = 42)
plot(v$avgJunctions, type = "l", xlab = "Generations",
ylab = "Number of Junctions", main = "Example Infinite Chromosome")
lines(v$detectedJunctions, col = "blue")
legend("bottomright", c("Real number","Number detected"),
lty = 1, col = c("black", "blue"))